Genus Alnus Mill. includes up to 40 species with a temperate Northern Hemisphere distribution (Komarov 1936; Davidov 1972; Furlow 1979; Tzvelev 2002). Among alder species, Alnus incana (L.) Moench. is the most widespread, and is widely known to be morphologically variable.
Some A. incana populations are so different from “typical” forms that various authors assign them the rank of species. One example is the “Kola alder” – Alnus kolaensis Orlova (1954) grows in Norway, Finland and Russia (on Kola Peninsula). Taxonomic status of this species is a matter of controversy. Orlova (1954), Ramenskaja and Andreeva (1982) accept it as a separate species, while Skvortzov (1959) and Nilsson (2000) believe it to be subspecies of A. incana, other authors recognize it as a hybrid between A. glutinosa (L.) Gaertn. and A. incana (Tzvelev, 2002), or as A. incana ecological form (Ilinsky, Shipunov, 2005), and some authors (Walters, 1993; Sokoloff & Filin, 1996) do not accept Kola alder as a taxon. The “true” (and usually sterile) hybrids between A. glutinosa and A. incana have been also described under the name Alnus × pubescens Taush (e.g., Nilsson, 2000).
With the purpose of clarifying taxonomical status of A. kolaensis, we performed genetic and morphometric analyses of four dominant alder species from the central and north-western regions of European Russia: Alnus incana, Alnus glutinosa, Alnus kolaensis and Alnus × pubescens.
We compared DNA sequences at the nuclear ITS locus. Such conservative site was selected because of risk to occur population heterogeneity described in literature (King & Ferris, 2000; Gomory & Paule, 2002). The lower heterogeneity of nuclear genes (in comparison with chloroplast genes) can help us show the gene flow, and usually each small difference is significant (Huh, 1999). ITS was also selected because the sequences for the greatest number of Alnus representatives, including A. incana and A. glutinosa are available from GenBank. The ITS site is often used in describing new species (Navarro et. al., 2004) and it is less conservative than other genes (like rbcL or 18S rRNA) used for similar purposes (Sanvard et al., 1993). Furthermore, as a nuclear region, ITS allow visualization of polyploidy in studied species (Jarvinen et. al., 2004). As A. kolaensis ITS sequences were absent from public databases such as GenBank, we sequenced the ITS locus for this taxon along with A. incana and A. glutinosa samples to approve database sequences.
For morphometric analysis, we have used classical and geometric techniques, because the combination of these methods provides the most complete view of similarities and distinctions among species (Ilinsky, Shipunov, 2005). Moreover, geometric morphometry is a tried-and-true method for complex group analyses, including taxa with hybrid origins (Jensen et al. 2002; Shipunov, Bateman 2005).
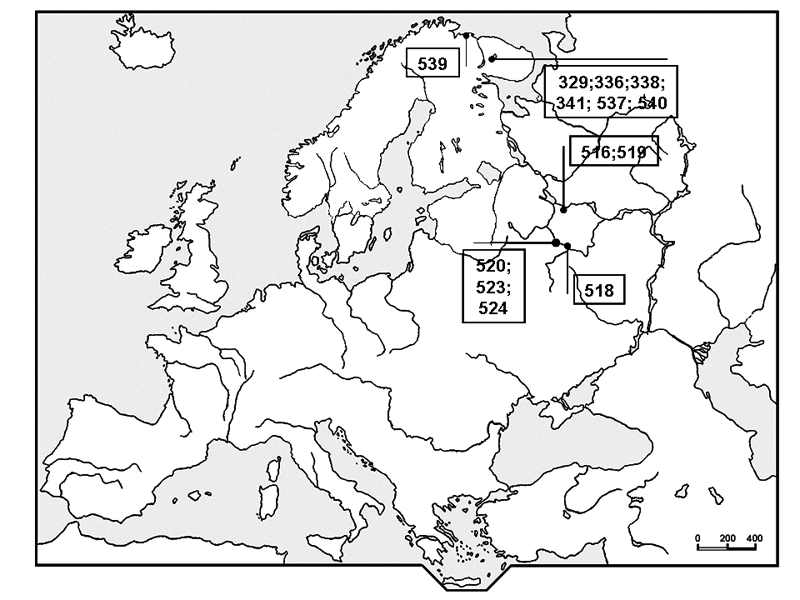
We used material from the Moscow State University Herbarium (MW) and the Botanical Institute of the Russian Academy of Science (LE) (Fig. 1). We also used morphological data from available literature (see Table 1). In all, 13 samples were sequenced.
Figure 1. A map of studied herbarium samples locations. Numbers are local database numbers (Table 1)

Table 1. Examined samples (ZIP, DOC inside)
As standard techniques of nuclear DNA extraction failed with herbarium material, a technique combining the classical CTAB method (Doyle JJ, Doyle JL, 1987) and a method, based on absorption with silicate carriers, has been used. First, DNA was extracted using classical CTAB method, and then it was cleaned using Probe-GS DNA Isolating Kit (DNA Technology, Inc.). Sequences at the ITS locus were obtained using universal primers (Aln-d 5' GAACCTGCGGAAGGATCATTGTC 3' and Aln-r 5' CGTTGCCGAGAGTCGTTATGGT 3') yielding a PCR product 256 bp length.
PCR conditions were 35 cycles of 94°C for 10 sec; 64°C for 10 sec and 72°C for 10 sec using a Tercyc PCR Cycler (DNA Technology, Inc.). Forward and reverse sequences were run using an ABI 3100 Avant with BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). LaserGene 4.0 (DNASTAR, Inc.) and Oligo 6.31 (Molecular Biology Insights, Inc.) were used for sequencing analysis.
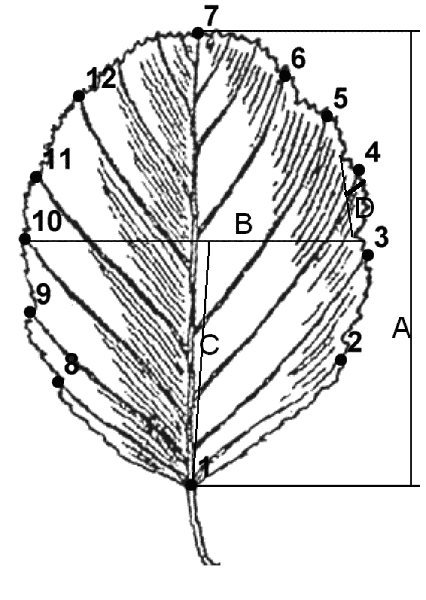
We performed morphometric analysis of leaf blade using geometric and classical morphometry methods, as previously described (Ilinsky, Shipunov, 2005). In the former, we used the technique with an arrangement of 12 landmarks on ends of first order veins (Fig. 2). Leaf parameters were chosen because other characteristics (e.g. mature stems, nutlet etc.) are quite similar among examined species (see Table 2). TPS program series and R 2.2.0 (R development core team, 2004) were used for morphometric analysis.
Figure 2. A scheme of landmarks location and measured parameters. Numbers show the order of landmarks. Letters show the following measurements: A – maximum leaf length, B – maximum leaf width, C – location of maximum width, D – length of maximum first order tooth
All sequences for A. incana and A. glutinosa coincided with sequences from NCBI GenBank. The sequences of five A. kolaensis samples were identical and differed both from A. incana and A. glutinosa by one nucleotide for each species. One herbarium sample identified as A. kolaensis represented a heterozygote on the investigated site; one allele corresponded to the A. kolaensis sequence, and another to A. incana. It has ovate leaves with an obtuse top are 60 mm in length and 40 mm in width, seven veins of the first order, hairy only on veins (see Fig. 4). Another herbarium sample identified as A. kolaensis matched the A. incana sequence at ITS locus.
Principal component analysis of both classical and geometric morphometry data shows high polymorphism among the alder groups in this study. Nevertheless, on the plot of first two components (Fig. 3) three sectors corresponding to A. glutinosa, A. incana and A. kolaensis sequences are well enough allocated. Prospective hybrid A. incana × A. kolaensis appears to take a marginal position within A. kolaensis sector. The A. ×pubescens sample is located separately from other three sectors.
Figure 3. The plot the first and the second components received from combination of classical and geometric morphometry methods. Letters indicate the following species: g – Alnus glutinosa, i – A. incana, k – A. kolaensis, p – A. × pubescens. Numbers indicate genotypes (ITS sequences): “-” – unknown, 1 – A. glutinosa, 2 – A. incana, 3 – A. kolaensis, 4 – A. incana × A. kolaensis
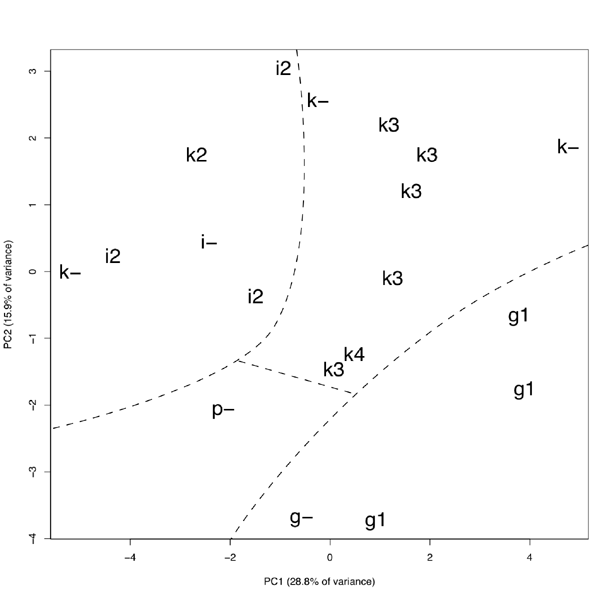
Figure 4. The heterozygous (Alnus incana × A. kolaensis) herbarium sample: whole view and a catkin

A. kolaensis and A. incana ITS1 sequences differ by one base pair, and due to low polymorphism amount studied species the building of polygenetic trees at this locus is not meaningful. However, these differences could be significant in the species’ taxonomic status (Chen & Li, 2004). In light of this high conservation at the ITS1 locus this data suggests that A. kolaensis should be accepted as a separate species (for reference, differences between Betula pendula Roth. and B. pubescens Erhr. are also one bp).
Heterozygous sample is apparently an occasional hybrid A. incana × A. kolaensis. Nevertheless, we cannot exclude, that A. incana allele is not segregating at a low frequency also in A. kolaensis, either because of past hybridization between the two species groups or due to shared polymorphism trough common ancestry.
According to our morphometric data, each sequence type (“taxon”) forms a separate sector, more or less isolated from the others, and A. ×pubescens hybrids take an intermediate position between their supposed parents. Morphometric data agree closely with molecular. One sample, marked in the herbarium as A. kolaensis, have A. incana genotype; moreover, it is located inside A. incana sector, as you can see on the graph (Fig. 3). Based on these data, we regard it as an incorrectly identified as an A. incana sample. One of O. Nilsson’s “A. incana subsp. kolaensis” samples is also located inside A. incana sector. Therefore, it seems that this subspecies is heterogeneous and it is better to go with N. Orlova (1954) diagnosis.
This all confirms that A. kolaensis merits taxonomic species status. In addition to DNA differences, these samples have distinct morphology: recursive tree analysis returned that the most significant characters are leaf height and width (less than 57.5 and 49 mm, respectively) and number of veins (less than 7). This description corresponds well with N.Orlova (1954) initial diagnosis.
Both morphological and sequence analysis yielded similar results, and it confirms previously-obtained findings from studies of Betulaceae (Bousquet et. al., 1992). Thus, we can use amount of morphological characteristics of the leaf blade and nuclear gene sequence in the systematics of the Betulaceae.
To our knowledge, the hybrid origin hypothesis of Alnus kolaensis has not been confirmed. Additionally, due to DNA differences within Kola alder, we cannot consider it as an ecological form. Other hypotheses regarding the origin of Alnus kolaensis remain unclear. However, molecular and morphological data show similar differences of Kola alder from its presumed parents, Alnus glutinosa and A. incana. Therefore, this taxon can be related to any of the examined species. Based on morphometric and genetic data, A. kolaensis must be considered an independent species, because the differences among other species show similar levels of difference. This conclusion will influence not only Alnus taxonomy but also the conservation policy in Murmansk region where many habitats of Kola alder are now under the effect of high air pollution (Zvereva & Kozlov, 2007).
We thank I.A. Kofiadi for help in DNA preparation and sequencing; collaborators of DNA-Technology JSC for given opportunities; participants and heads of Moscow South-West High School 1543 expeditions; and collaborators of MSU and BIN RAS herbariums for assistance rendered. We also heartily thank A.-J. Brumble for reviewing the English translation of initial manuscript.